How can I speed up my R code? Part 1
I’ve had relatively few experiences in using R so far which ran frustratingly slowly. But when they did run slowly, it was excruciatingly slow and frustrating to watch. I’ve heard time and time again that “R is slow” and that I should exepect/deal with it or move to a) a more complicated language like C/C++ or b) a more expensive stats programme.
But the truth is there’s a good number of ways making R code run faster, and in almost every case probably as fast as you need it to! Here are a few tips I’ve come across on how to speed things up – with hopefully some insights into how R works behind the scenes!
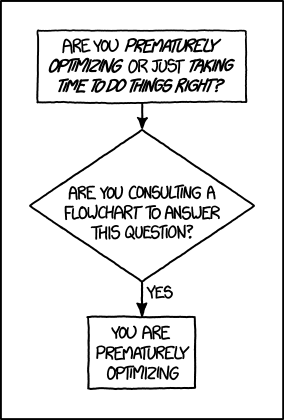
Before you start optimising – Don’t!

Before you start optiming your code, STOP! And think about whether you really need to do this! There’s a thing called “Premature Optimisation” where you get too bogged down in the details of how to speed something up, whent it doesn’t really matter!
If you’re running code, and it takes a few seconds, how much does it really matter if you can shave off half a second? Especially if you’re a researcher like me and only need to run it a couple of times before it’s archived and you move on.
You could end up wasting a lot more time just trying to figure out techniques for optimising and speeding up your code! So before you even get going with that, have a good think about whether you need to.
Having said that, it is more fun to figure out complex problems! And if you’re a PhD student or a researcher in training, then spending some time gaining a Transferrable Skill such as optimisation might well be worth it! Right??
Additionally, there are some things you can do really easily to make your code better. So read on and I’ll talk through some ideas, from simple changes of habit up to complex tools you can use.
Problem 1 – Bad code
Or: why for loops are awful and you should never use them
What’s slowing things down?
Bad code is a common problem, and likely comes from learning bad habits through awful intro to R courses or through transfers from other languages. The use of ‘for’ loops is a common bad habit that you should unlearn as a priority!
A helpful first step is uUnderstanding how R works, what its limitations are, and what’s built into it already to overcome these limitations.
You see, R has actually some really fast bits under the hood, written in C (a compiled language). This means that when R is given a simple instruction it can work out how to do it really quickly.
The problem with for loops is it essentially breaks up the cycle of quick parts by turning back to slow parts.
Imagine for a moment you run a newsagents, and you have a kid on a bike who’s tasked with delivering papers to the neighbourhood. In the shop on a clipboard you have a vector of house numbers and a vector of papers to deliver the houses to.
‘Aha!’ you say to yourself, ‘It’s time for a for loop!’. You iterate through that list, counting up the number of houses, finding the correct paper and passing both address and paper to the paperboy (who’s actually a very fast cyclist). When he returns and reports success, you increment your counter by one, find the next address and paper, and depart him on his way again:
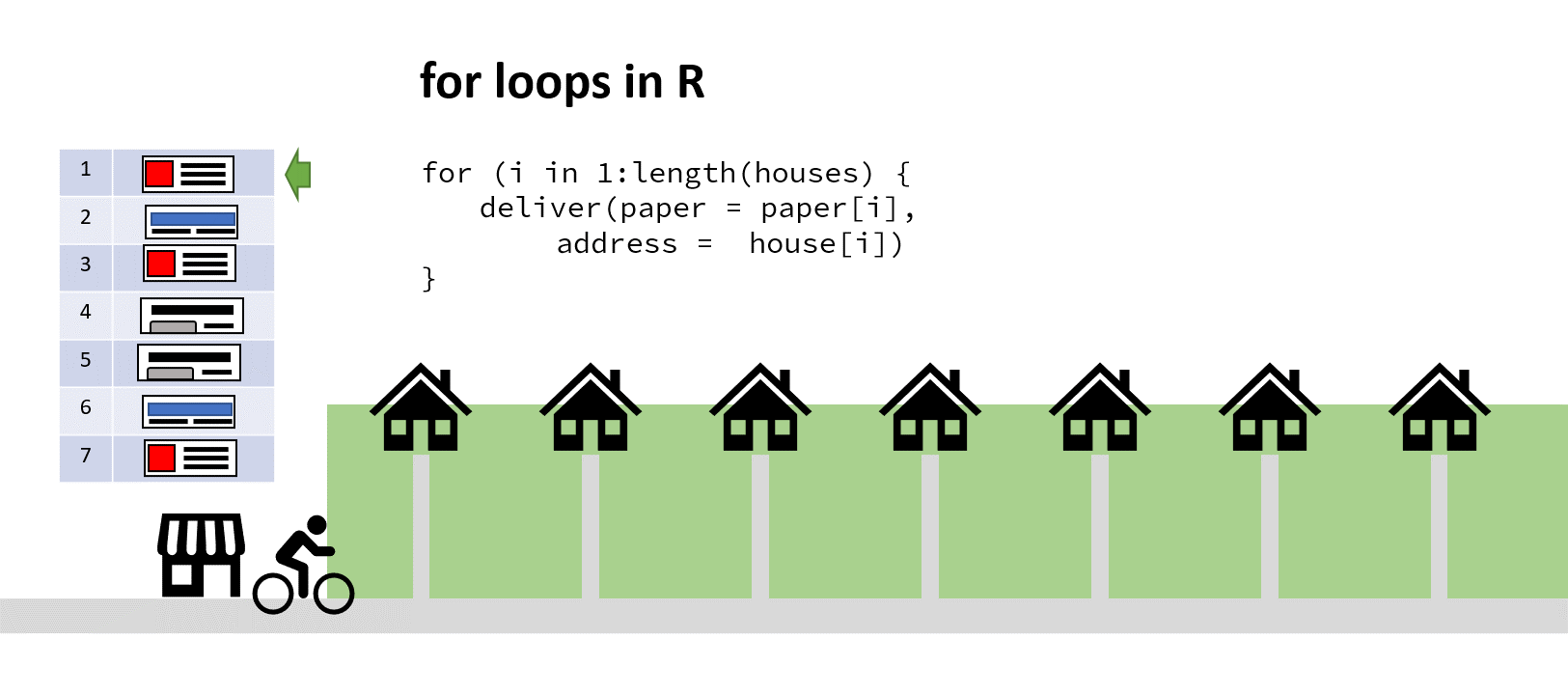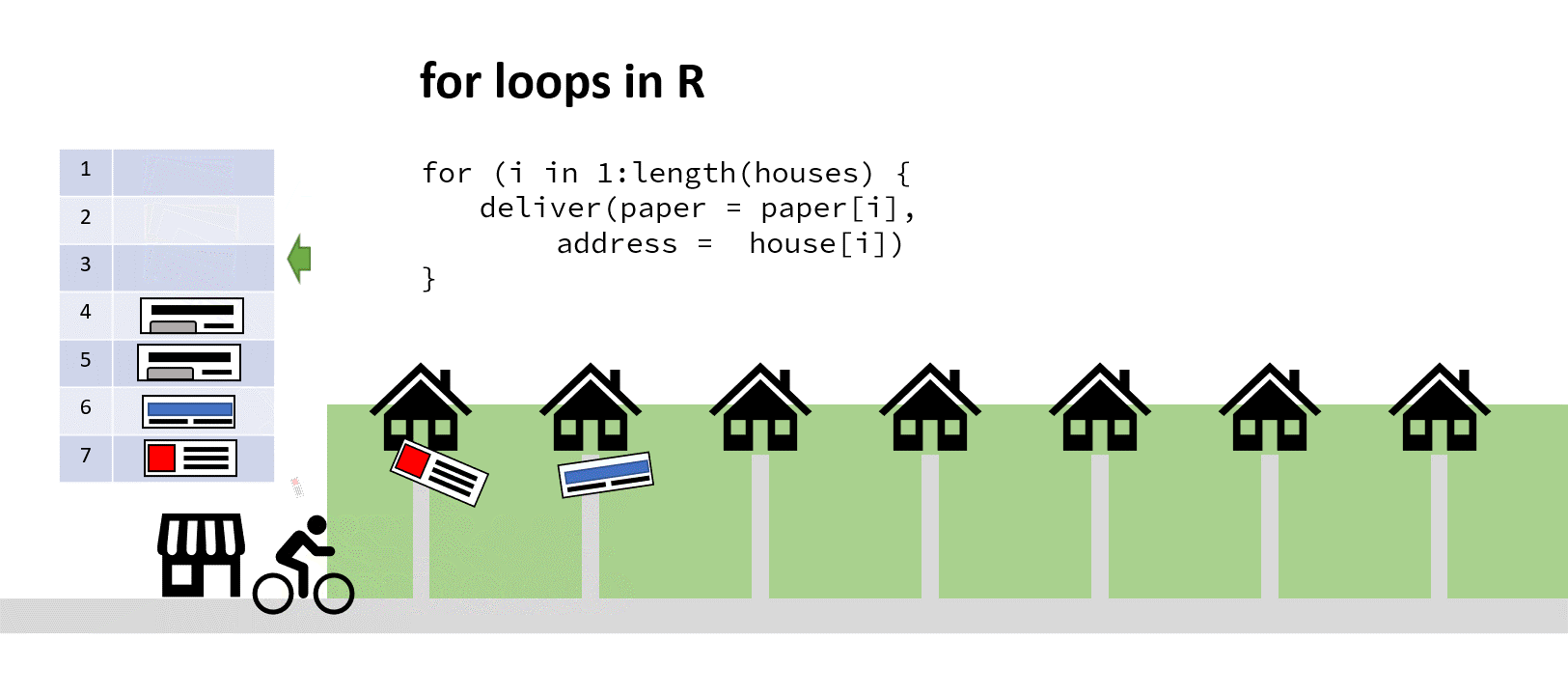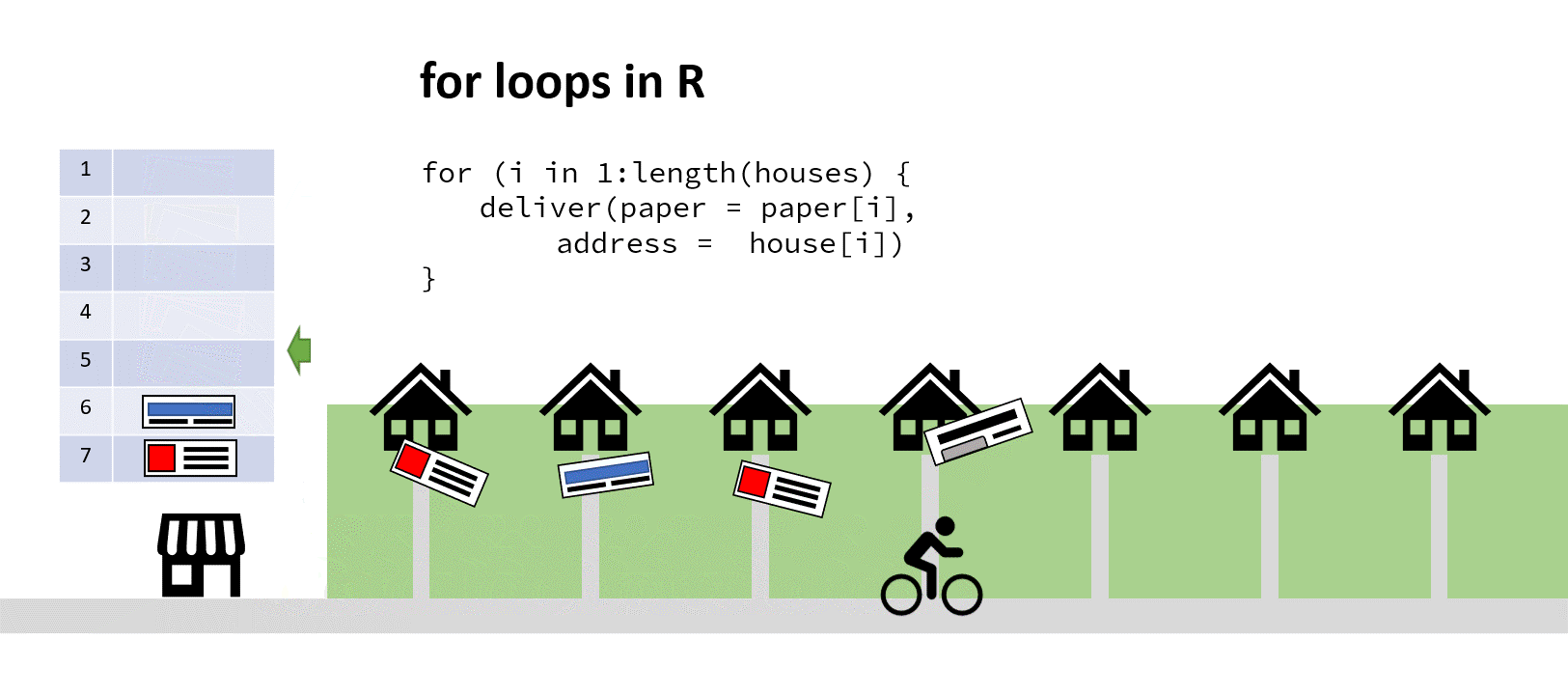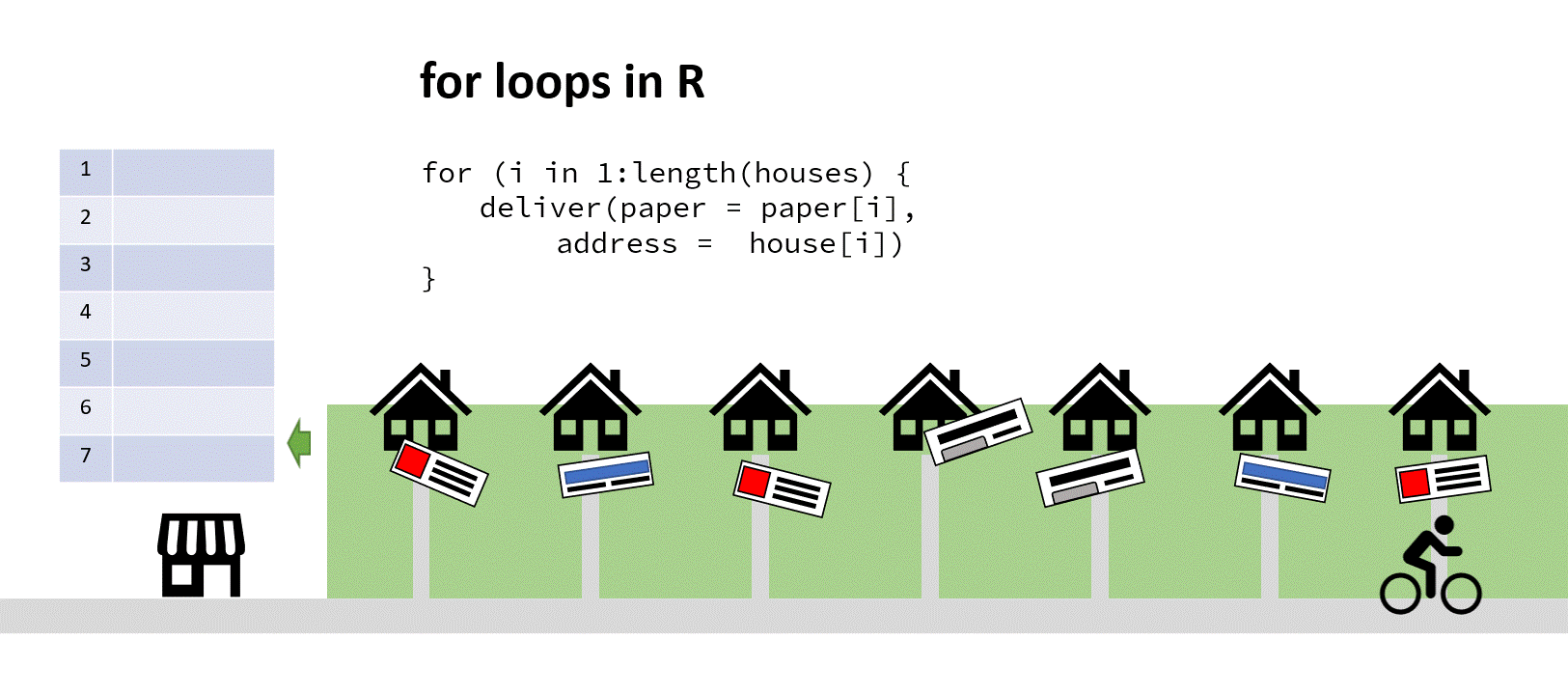 The cyclist here saves time by not turning around (my animation skills got lazy at this point)
The cyclist here saves time by not turning around (my animation skills got lazy at this point)
Now, pause a second here and try to think of a better way of doing this…
…
You’ve hopefully come up with the same solution here! Assuming a sack large enough, you could pass the paperboy all the papers to iterate through, and all the addresses to simultaneously iterate through. Then he wouldn’t have to keep returning to the shop to find out the next paper/address combo:
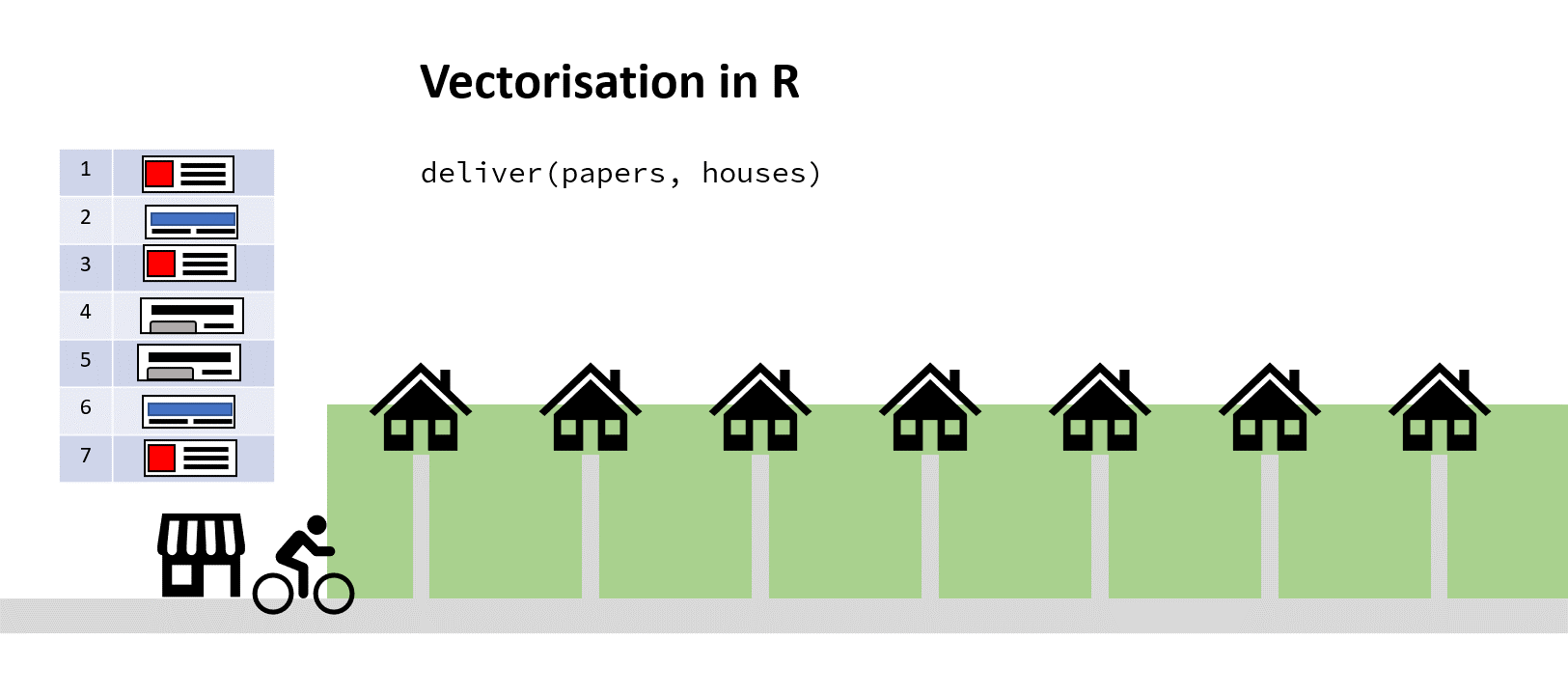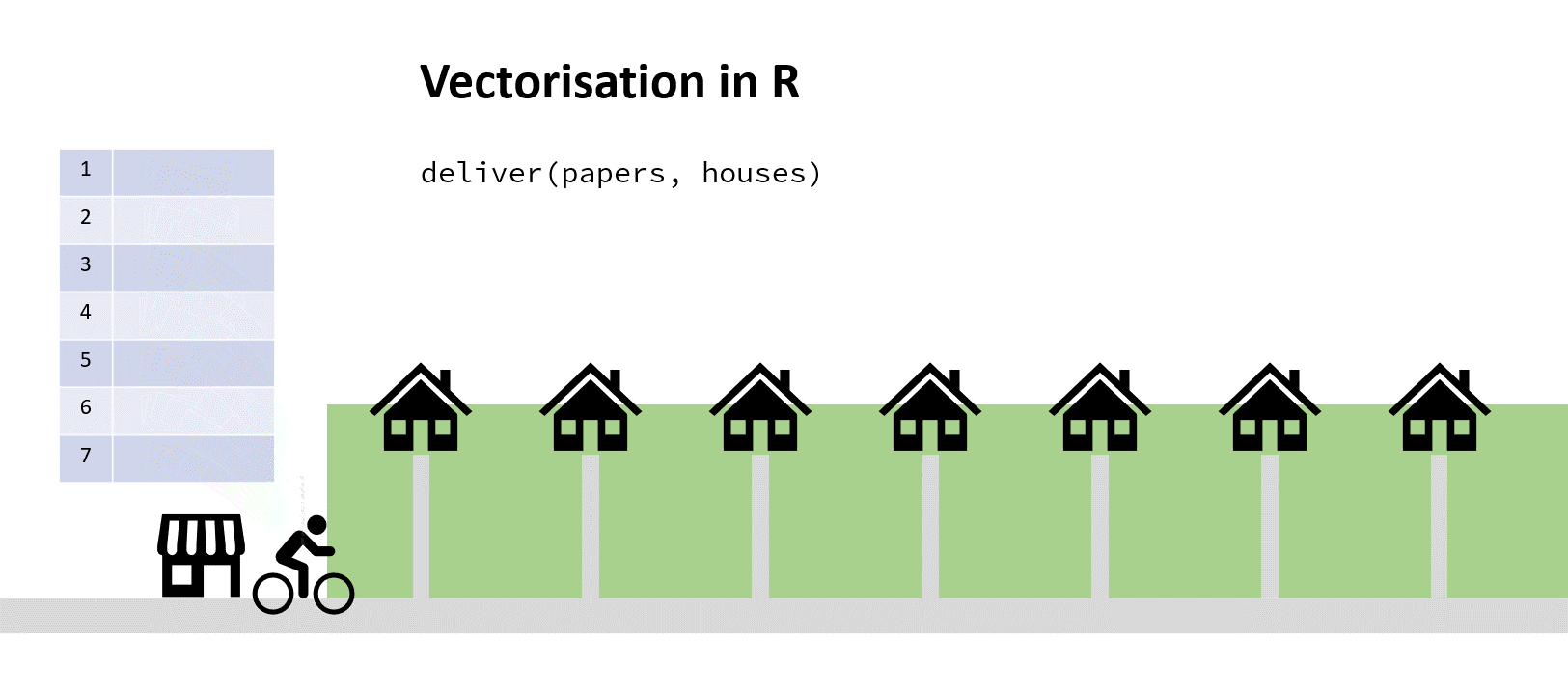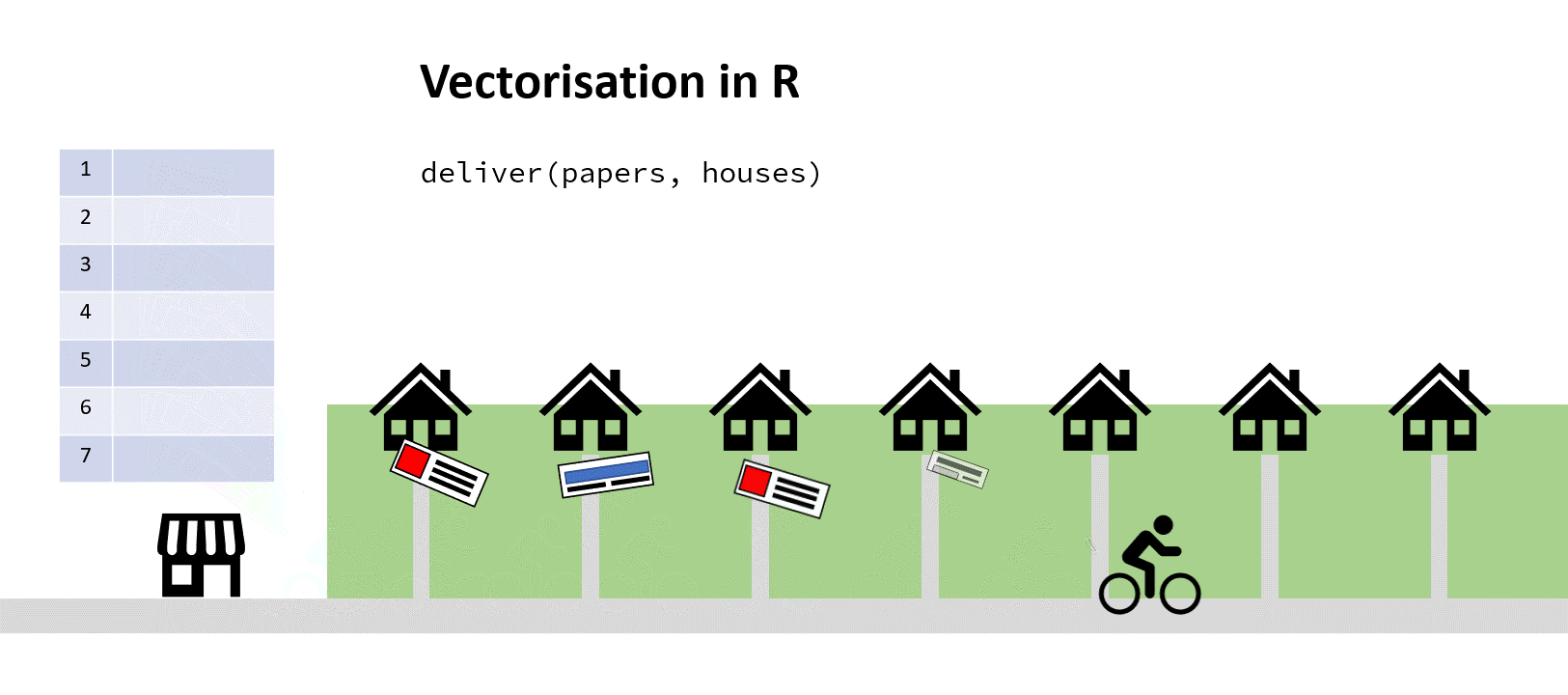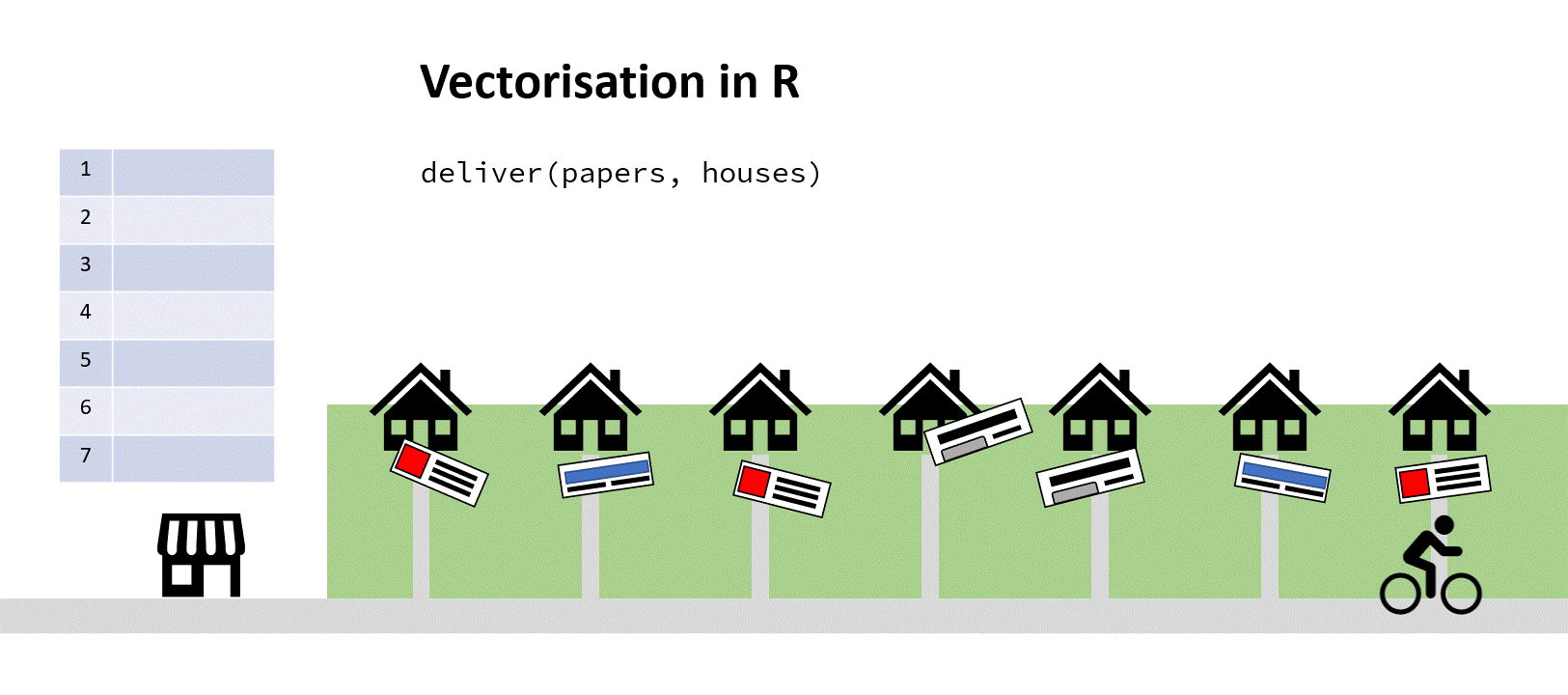
Solution 1 – Vectorisation
This is essentially (with a bit of poetic license) what is going on when you use what’s called vectorisation in R instead of using a for loop.
In R it’s the difference between doing:
nums <- 1:10
total <- 0
for (i in 1:length(nums)) {
total <- total + nums[i]
}
and
total <- sum(nums)
When you get to big numbers and large dataframes, the difference in timings can be quite substantial! (pro tip: “A data frame is a named list of vectors”) For loops in general can and should always be replaced by passing R your whole vector (or dataframe column, or list of objects) at once to perform the whole function on.
Lots of functions in R are strictly vectorised – as in, they are programmed to do things really fast to vectors. These include the base aritmetic functions (+, -, /, *, etc.), logical functions (&, |, etc.) and a bunch of other handy summarising functions (e.g. sum(), rowsums()).
These run faster, look tider and are easier to read and debug than for loops!
Even without pure vectorised functions, there are still very good reasons to use codes which deals with whole vectors/columns/lists at the same time. This is where functionals come in handy! Learning to use base R’s apply family or purrr’s map functions can save a lot of time, confusion and untidiness.
Using vectors/functionals to speed up (and tidy up) your R code by replacing for loops is a very simple time-saving tip that you should take on as a new habit.
A worked example
To show how this works, and how you might have to put a bit of thought into using it, here’s an example from a StackOverflow question I came across a while ago.
The question centered around how to update a value in a row using a calculation from a previous row. The example equation was simple, multiply the last value by 0.2. The questioner used a for-loop for this, but with a large dataset it was taking hours, if not months to do!
Here’s a for-loop implementation over 100,000 rows:
library(tidyverse)
df <- tibble(row = 1:100000,
A = c(1, rep(NA, 99999)))
t1 <- Sys.time()
for (i in 2:nrow(df)) {
df$A[i] <- 0.2 * df$A[i-1]
}
cat("That took ", Sys.time() - t1, "seconds")
#> That took 47.32143 seconds
47 seconds is a pain! So can it be done faster with vectorisation?
The big problem here is how can you pass R the whole vector at once for one calculation? Each calculation depends on the previous one being calculated. We cannot tell the value of \(A_n\) without first calculating \(A_{(n-1)}\). Or can we?
The neat thing to notice here is that each row has its sequential row number: row, and the calculation that depends on the previous value is actually just multiplying the base value by the 0.2 coefficient (row - 1) times:
This means that in reality R can calculate all values ‘at once’ by passing it a neater equation (after calculating a base A value first):
library(tidyverse)
df <- tibble(row = 1:100000,
A = c(1, rep(NA, 99999)))
t1 <- Sys.time()
df <- df |>
mutate(A = max(A, na.rm = TRUE) * 0.2 ^ (row - 1))
cat("That took ", Sys.time() - t1, "seconds")
#> That took 0.07244086 seconds
That did exactly the same thing in mere fragments of the original time! Pretty cool huh?
This is what I mean by leveraging very simple habits and tweaks to your code to get things running faster. It uses a few simple principles:
- Don’t use
forloops - Use ‘vectorised’ functions where you can
- Pass whole vectors as arguments when a function can handle them
- Use functionals (
map/lapply, etc.) when it cant
But can speed up (and tidy up!) your code substantially in the long run.
That still not super-speeded up your data though? Perhaps you’ve got a ‘Problem 2 – Big data’. Well stay tuned for Part 2 of speeding up your R code!